Tutorial 1: Built-in demonstration scripts¶
MAVE-NN provides built-in demonstration scripts, or “demos”, to help users quickly get started training and visualizing models. Demos are self-contained Python scripts that can be executed by calling mavenn.run_demo. To get a list of demo names, execute this function without passing any arguments:
[1]:
# Import MAVE-NN
import mavenn
# Get list of demos
mavenn.run_demo()
To run a demo, execute
>>> mavenn.run_demo(name)
where 'name' is one of the following strings:
1. "gb1_ge_evaluation"
2. "mpsa_ge_training"
3. "sortseq_mpa_visualization"
Python code for each demo is located in
/Users/jkinney/github/mavenn/mavenn/examples/demos/
[1]:
['gb1_ge_evaluation', 'mpsa_ge_training', 'sortseq_mpa_visualization']
To see the Python code for any one of these demos, pass the keyword argument print_code=True to mavenn.run_demo(). Alternatively, navigate to the folder that is printed when executing mavenn.run_demo() on your machine and open the corresponding *.py file.
Evaluating a GE regression model¶
The 'gb1_ge_evaluation' demo illustrates an additive G-P map and GE measurement process fit to data from a deep mutational scanning (DMS) experiment performed on protein GB1 by Olson et al., 2014.
[2]:
mavenn.run_demo('gb1_ge_evaluation', print_code=False)
Running /Users/jkinney/github/mavenn/mavenn/examples/demos/gb1_ge_evaluation.py...
Using mavenn at: /Users/jkinney/github/mavenn/mavenn
Model loaded from these files:
/Users/jkinney/github/mavenn/mavenn/examples/models/gb1_ge_additive.pickle
/Users/jkinney/github/mavenn/mavenn/examples/models/gb1_ge_additive.h5
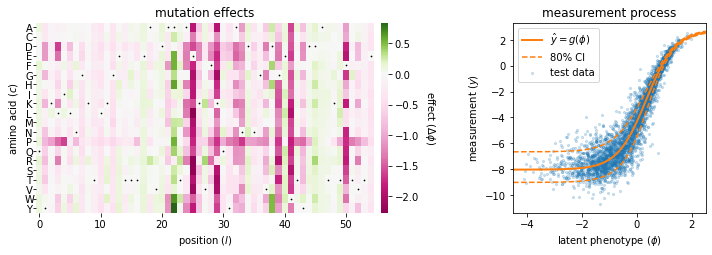
Done!
Visualizing an MPA regression model¶
The 'sortseq_mpa_visualization' demo illustrates an additive G-P map, along with an MPA measurement process, fit to data from a sort-seq MPRA performed by Kinney et al., 2010.
[3]:
mavenn.run_demo('sortseq_mpa_visualization', print_code=False)
Running /Users/jkinney/github/mavenn/mavenn/examples/demos/sortseq_mpa_visualization.py...
Using mavenn at: /Users/jkinney/github/mavenn/mavenn
Model loaded from these files:
/Users/jkinney/github/mavenn/mavenn/examples/models/sortseq_full-wt_mpa_additive.pickle
/Users/jkinney/github/mavenn/mavenn/examples/models/sortseq_full-wt_mpa_additive.h5
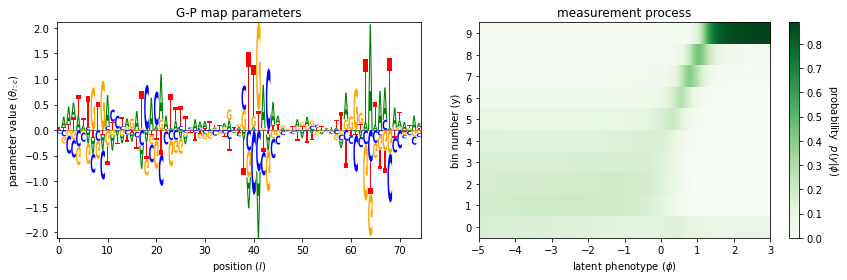
Done!
Training a GE regression model¶
The 'mpsa_ge_training' demo uses GE regression to train a pairwise G-P map on data from a massively parallel splicing assay (MPSA) reported by Wong et al., 2018. This training process usually takes under a minute on a standard laptop.
[4]:
mavenn.run_demo('mpsa_ge_training', print_code=False)
Running /Users/jkinney/github/mavenn/mavenn/examples/demos/mpsa_ge_training.py...
Using mavenn at: /Users/jkinney/github/mavenn/mavenn
N = 24,405 observations set as training data.
Using 19.9% for validation.
Data shuffled.
Time to set data: 0.208 sec.
LSMR Least-squares solution of Ax = b
The matrix A has 19540 rows and 36 columns
damp = 0.00000000000000e+00
atol = 1.00e-06 conlim = 1.00e+08
btol = 1.00e-06 maxiter = 36
itn x(1) norm r norm Ar compatible LS norm A cond A
0 0.00000e+00 1.391e+02 4.673e+03 1.0e+00 2.4e-01
1 2.09956e-04 1.273e+02 2.143e+03 9.2e-01 2.1e-01 8.1e+01 1.0e+00
2 4.17536e-03 1.263e+02 1.369e+03 9.1e-01 5.1e-02 2.1e+02 1.9e+00
3 3.65731e-03 1.251e+02 5.501e+01 9.0e-01 1.6e-03 2.8e+02 2.6e+00
4 3.27390e-03 1.251e+02 6.185e+00 9.0e-01 1.7e-04 2.9e+02 3.2e+00
5 3.20902e-03 1.251e+02 3.988e-01 9.0e-01 1.1e-05 3.0e+02 3.4e+00
6 3.21716e-03 1.251e+02 1.126e-02 9.0e-01 3.0e-07 3.0e+02 3.4e+00
LSMR finished
The least-squares solution is good enough, given atol
istop = 2 normr = 1.3e+02
normA = 3.0e+02 normAr = 1.1e-02
itn = 6 condA = 3.4e+00
normx = 8.2e-01
6 3.21716e-03 1.251e+02 1.126e-02
9.0e-01 3.0e-07 3.0e+02 3.4e+00
Linear regression time: 0.0044 sec
Epoch 1/30
391/391 [==============================] - 1s 1ms/step - loss: 48.0562 - I_var: 0.0115 - val_loss: 41.2422 - val_I_var: 0.1856
Epoch 2/30
391/391 [==============================] - 0s 686us/step - loss: 41.0110 - I_var: 0.1912 - val_loss: 40.7058 - val_I_var: 0.1993
Epoch 3/30
391/391 [==============================] - 0s 682us/step - loss: 40.3925 - I_var: 0.2076 - val_loss: 40.0481 - val_I_var: 0.2178
Epoch 4/30
391/391 [==============================] - 0s 684us/step - loss: 39.9577 - I_var: 0.2203 - val_loss: 40.1940 - val_I_var: 0.2132
Epoch 5/30
391/391 [==============================] - 0s 687us/step - loss: 39.7409 - I_var: 0.2264 - val_loss: 39.8678 - val_I_var: 0.2234
Epoch 6/30
391/391 [==============================] - 0s 690us/step - loss: 39.4834 - I_var: 0.2343 - val_loss: 39.4798 - val_I_var: 0.2351
Epoch 7/30
391/391 [==============================] - 0s 688us/step - loss: 39.3679 - I_var: 0.2381 - val_loss: 39.6919 - val_I_var: 0.2293
Epoch 8/30
391/391 [==============================] - 0s 688us/step - loss: 39.2042 - I_var: 0.2433 - val_loss: 39.1096 - val_I_var: 0.2462
Epoch 9/30
391/391 [==============================] - 0s 691us/step - loss: 39.1603 - I_var: 0.2452 - val_loss: 38.8807 - val_I_var: 0.2537
Epoch 10/30
391/391 [==============================] - 0s 701us/step - loss: 38.9189 - I_var: 0.2529 - val_loss: 38.8879 - val_I_var: 0.2537
Epoch 11/30
391/391 [==============================] - 0s 709us/step - loss: 38.6567 - I_var: 0.2612 - val_loss: 38.4904 - val_I_var: 0.2662
Epoch 12/30
391/391 [==============================] - 0s 686us/step - loss: 38.5589 - I_var: 0.2648 - val_loss: 39.1242 - val_I_var: 0.2490
Epoch 13/30
391/391 [==============================] - 0s 682us/step - loss: 38.3520 - I_var: 0.2723 - val_loss: 37.8326 - val_I_var: 0.2877
Epoch 14/30
391/391 [==============================] - 0s 688us/step - loss: 37.7298 - I_var: 0.2920 - val_loss: 37.4434 - val_I_var: 0.3001
Epoch 15/30
391/391 [==============================] - 0s 690us/step - loss: 37.3966 - I_var: 0.3027 - val_loss: 36.5743 - val_I_var: 0.3259
Epoch 16/30
391/391 [==============================] - 0s 690us/step - loss: 36.7566 - I_var: 0.3217 - val_loss: 36.6817 - val_I_var: 0.3231
Epoch 17/30
391/391 [==============================] - 0s 707us/step - loss: 36.5558 - I_var: 0.3276 - val_loss: 37.0841 - val_I_var: 0.3117
Epoch 18/30
391/391 [==============================] - 0s 699us/step - loss: 36.4968 - I_var: 0.3290 - val_loss: 36.1766 - val_I_var: 0.3373
Epoch 19/30
391/391 [==============================] - 0s 691us/step - loss: 36.3217 - I_var: 0.3340 - val_loss: 36.4613 - val_I_var: 0.3290
Epoch 20/30
391/391 [==============================] - 0s 687us/step - loss: 36.3226 - I_var: 0.3337 - val_loss: 36.2868 - val_I_var: 0.3347
Epoch 21/30
391/391 [==============================] - 0s 702us/step - loss: 36.3051 - I_var: 0.3345 - val_loss: 36.2097 - val_I_var: 0.3359
Epoch 22/30
391/391 [==============================] - 0s 690us/step - loss: 36.2569 - I_var: 0.3356 - val_loss: 36.0617 - val_I_var: 0.3406
Epoch 23/30
391/391 [==============================] - 0s 688us/step - loss: 36.1654 - I_var: 0.3384 - val_loss: 37.0675 - val_I_var: 0.3118
Epoch 24/30
391/391 [==============================] - 0s 697us/step - loss: 36.1285 - I_var: 0.3395 - val_loss: 36.0666 - val_I_var: 0.3403
Epoch 25/30
391/391 [==============================] - 0s 694us/step - loss: 36.0856 - I_var: 0.3409 - val_loss: 36.4473 - val_I_var: 0.3296
Epoch 26/30
391/391 [==============================] - 0s 688us/step - loss: 36.0640 - I_var: 0.3416 - val_loss: 36.2272 - val_I_var: 0.3363
Epoch 27/30
391/391 [==============================] - 0s 699us/step - loss: 36.0285 - I_var: 0.3426 - val_loss: 36.2458 - val_I_var: 0.3356
Epoch 28/30
391/391 [==============================] - 0s 703us/step - loss: 36.2016 - I_var: 0.3377 - val_loss: 36.5967 - val_I_var: 0.3250
Epoch 29/30
391/391 [==============================] - 0s 689us/step - loss: 36.0952 - I_var: 0.3408 - val_loss: 36.1920 - val_I_var: 0.3369
Epoch 30/30
391/391 [==============================] - 0s 699us/step - loss: 36.0361 - I_var: 0.3427 - val_loss: 36.3018 - val_I_var: 0.3338
Training time: 9.0 seconds
I_var_test: 0.335 +- 0.024 bits
I_pred_test: 0.367 +- 0.016 bits
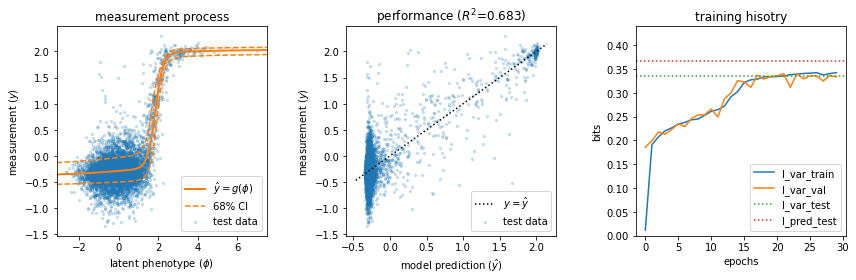
Done!
References¶
Kinney J, Murugan A, Callan C, Cox E. Using deep sequencing to characterize the biophysical mechanism of a transcriptional regulatory sequence. Proc Natl Acad Sci USA. 107:9158-9163 (2010).
Olson CA, Wu NC, Sun R. A comprehensive biophysical description of pairwise epistasis throughout an entire protein domain. Curr Biol 24:2643–2651 (2014).
Wong M, Kinney J, Krainer A. Quantitative activity profile and context dependence of all human 5’ splice sites. Mol Cell 71:1012-1026.e3 (2018).
[ ]: